It is thought that nearly half of infertility cases have a genetic basis. Despite extensive knowledge gained from gene knockouts in mice, the genetic causes for the vast majority of idiopathic human infertilities are unknown. Traditional methods for studying inheritance, such as GWAS or linkage analysis, have been confounded by heterogeneity of infertility phenotypes and hundreds of genes involved in reproduction. Finally, we do not know the proportion of cases in which genetically-based infertilities are caused by de novo mutations vs. inheritance of alleles segregating in the human population.
We are working on three separate funded projects to address the genetic and epigenetics of infertility.
The first project, funded by NICHD (R01HD082568), is entitled "Identification and validation of human infertility alleles.” This project employs a novel approach to this problem as outlined in the figure below. The Schimenti lab, in collaboration with the labs of Haiyuan Yu and Charles Danko at Cornell, are identifying nonsynonymous single nucleotide polymorphisms (nsSNPs; those that alter an amino acid) that functionally disrupt gametogenesis, purely by mining available human data on genetic variants. We are also identifying regulatory elements in the genome that are key for meiosis-expressed genes, and which contain variants that impact the function of those elements (thus impacting expression of the associated gene.
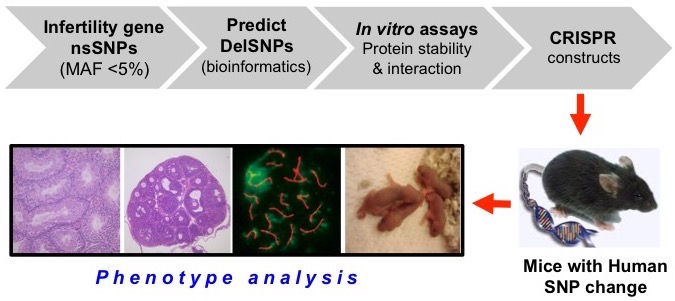
A second project is part of an NIH Center grant (P50HD096723) headed by Kyle Orwig at the University of Pittsburgh, and is a collaboration between Kyle Orwig, John Schimenti, Don Conrad, and Alex Yatsenko. The Schimenti lab’s role is to model candidate human variants identified by sequencing of male infertility patients, to determine if these variants are indeed underlying the phenotypes presented in those patients. We are also addressing potential epigenetic causes of infertility using CRISPRi and CRISPRa screening of several hundred epigenetic-related genes in PGCLCs (primordial germ cell-like cells) made in vitro.
The third major project is part of another NIH Center grant (P50HD104454) headed by Paula Cohen at Cornell. Dr. Schimenti’s project is in collaboration with Andrew Grimson (Cornell) and Kat Huang (Pittsburgh), and the goal is to identify variants in the 3’UTR of essential spermatogenesis genes that cause infertility. Suspect variants are being identified by sequencing of round spermatids from the ejaculates of sterile men, and also via bioinformatic identification of variants from the gnomAD database. We are emphasizing variants/mutations that impact miRNA binding sites and NRA binding protein binding sites.
List of SNPs tested via in vitro assays